Projects
-
Interpretable integration of multiomic data to study complex diseases: The DSPN study
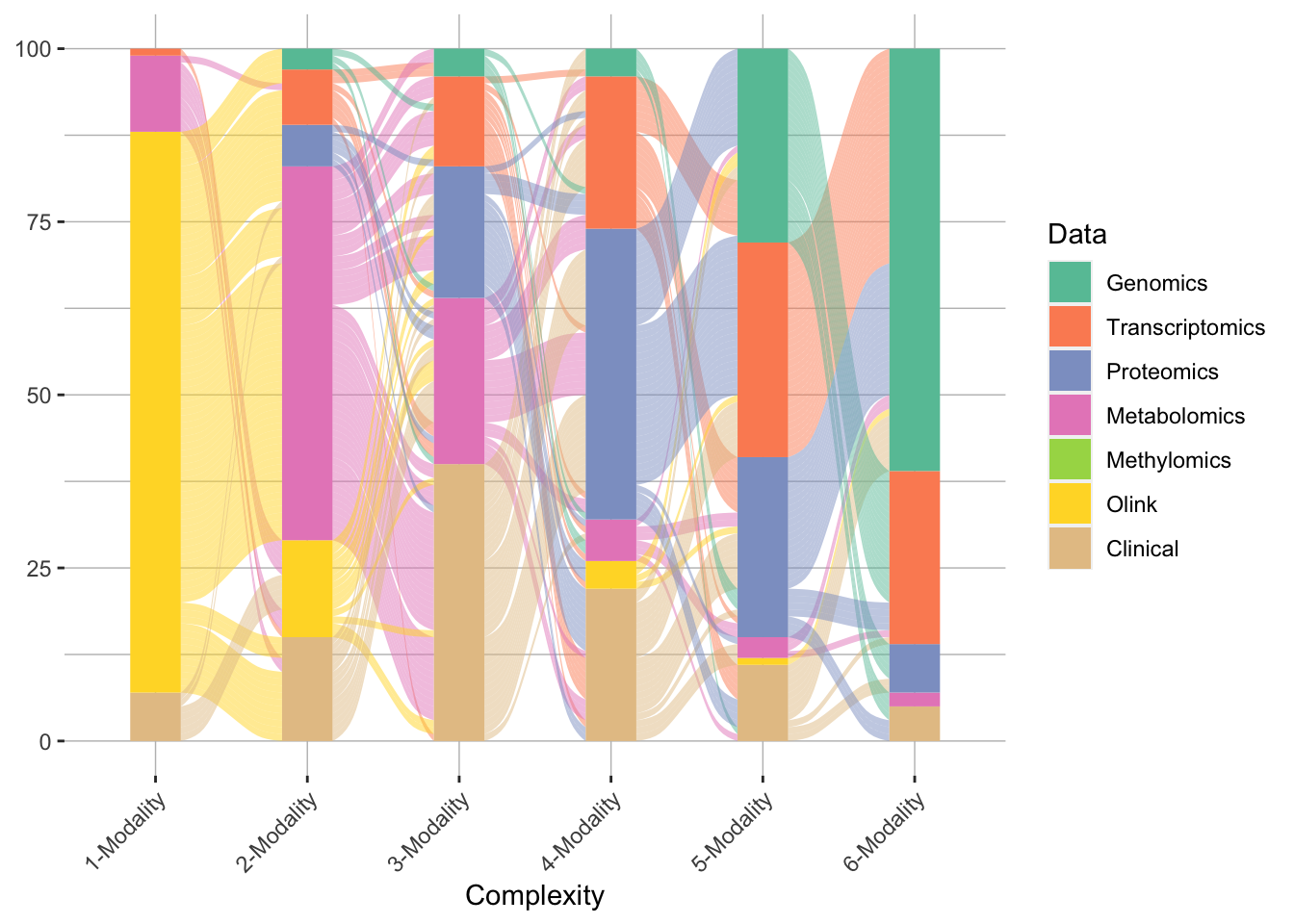
Complex diseases present a significant challenge to modern medicine due to their multifactorial nature. Understanding these diseases requires the integration of diverse datasets that encompass clinical, environmental, genetic and metabolic factors. In recent years, together with the advances of molecular characterization technologies, the emergence of large multiomic and clinical datasets has opened new opportunities to study such diseases comprehensively. In this article, I discuss the challenges of stdudying complex diseases using multimodal datasets, our approach to tackle these challenges and an example study to demonstrate the effectiveness of our method.
-
The Interpretable Multimodal Machine Learning (IMML) framework
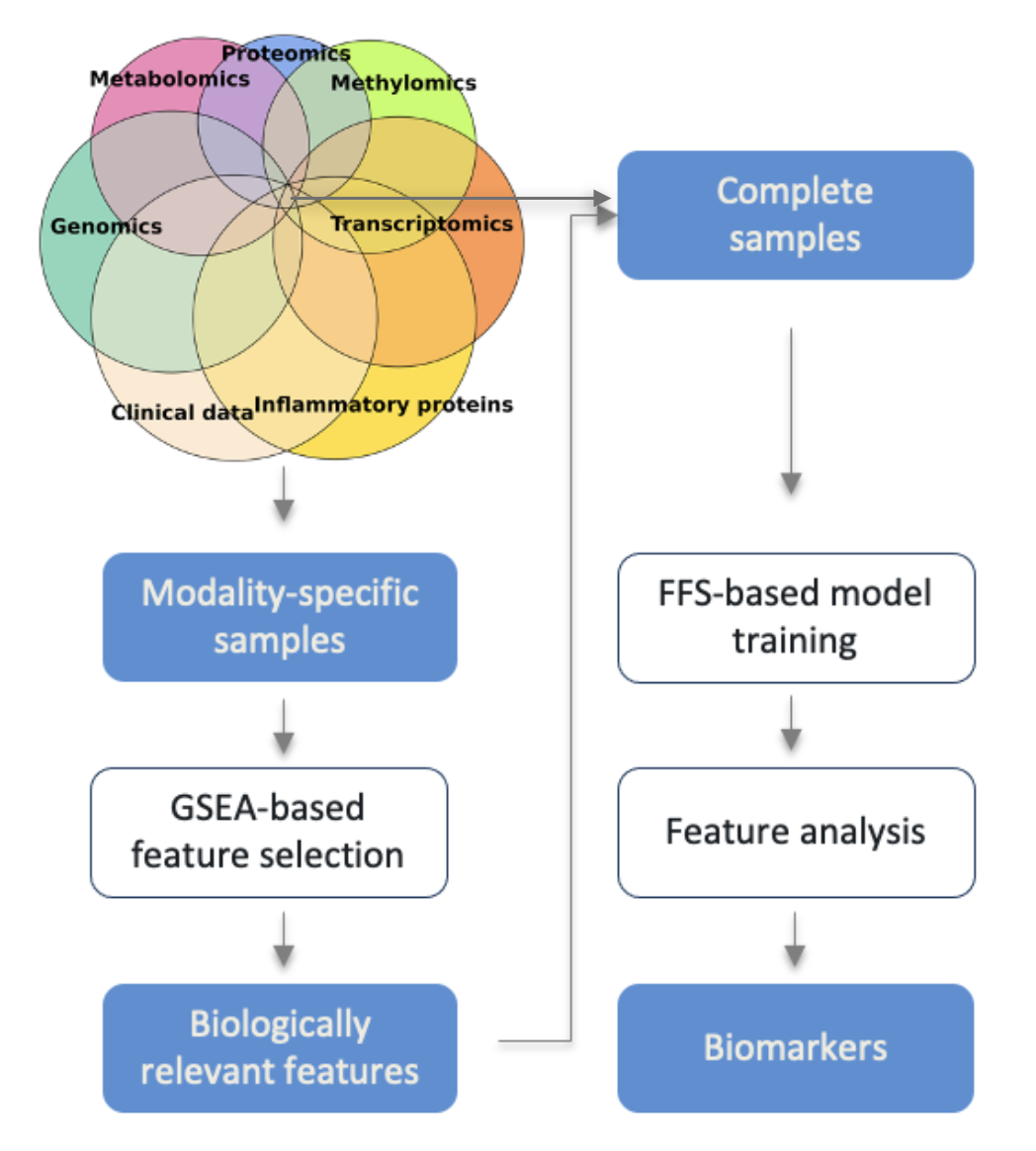
In the previous post we got to know the IMML and its application in the DSPN study. In this post I would like to go more into details of the algorithm, how it is constructed, what it actually does underneath and how it helps in studying complex deseases. This is more of a discription of the algorithm, rather than a coding tutorial. For a full on tutorial of how to actually use IMML, please visit my github repository.
-
Prognosis of type 2 diabetes leveraging pretrained large language model for longitudinal medical records
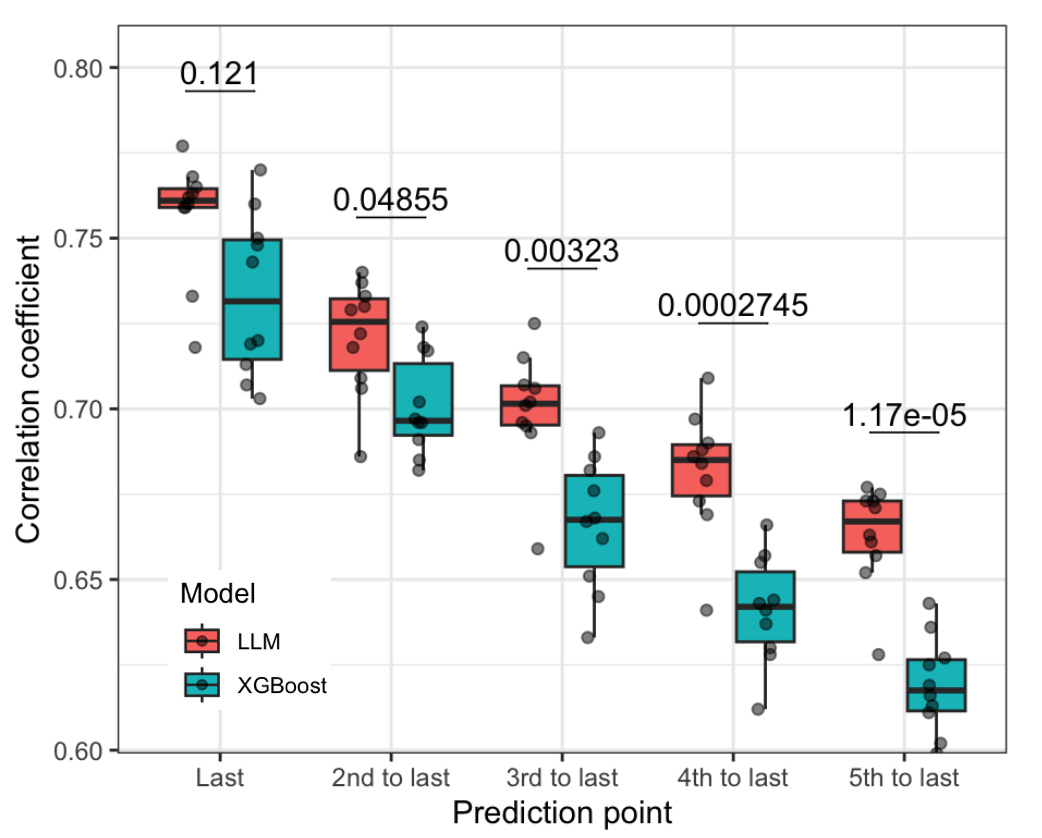
Type 2 diabetes (T2D) is a chronic metabolic disorder characterized by elevated blood glucose levels and is linked to severe microvascular and macrovascular complications such as cardiovascular diseases, neuropathy, nephropathy, and retinopathy, which significantly contribute to global morbidity and mortality. With T2D prevalence rising worldwide, effective management and early prognosis of its complications are essential for reducing healthcare costs and improving patient outcomes. This project aims to assist T2D leveraging availablity of large medical datasets and advancements in AI-driven large language models.
-
The adaptation of pretrained LLM to the medical domain
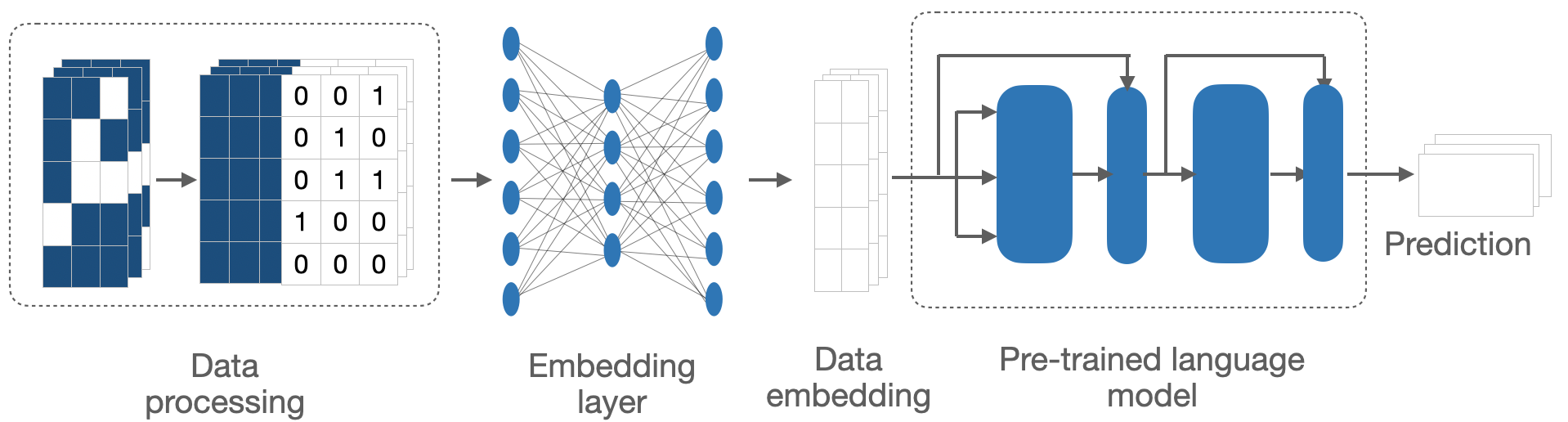
In this post, I aim to provide you with the technical details of how I adapted the natural language-based LLMs to longitudinal sparse numerical data of medical domain. Particularly, I will describe how I generated synthetic data to overcome the issues of data privacy and resource limitations; and the architecture of the adapted LLMs to bridge the gaps between the two domains. For details of applications and performance, please visit the previous post.
-
Evaluation of the impact of ancestry to drug response in cancer cell lines
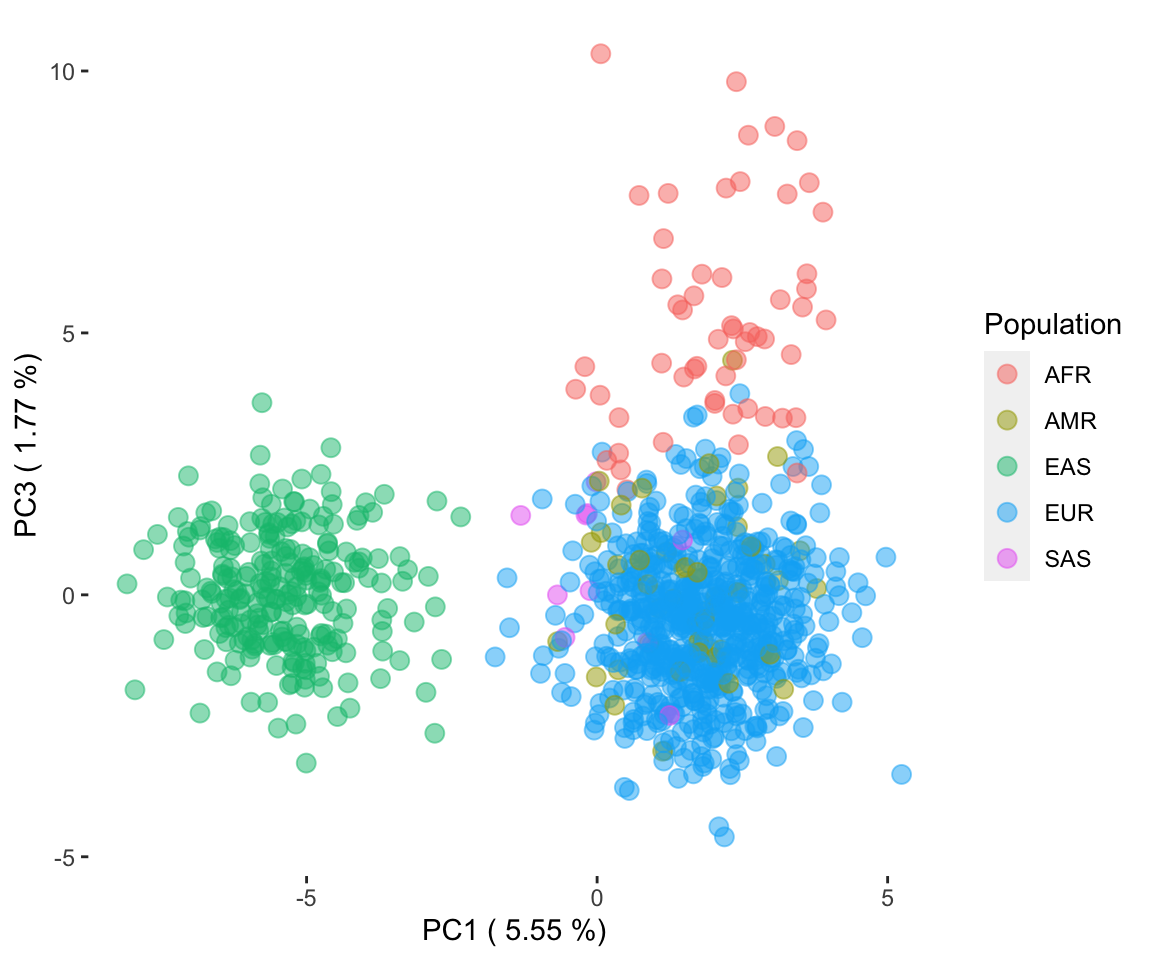
Cancer research has long relied on the use of in vitro cell lines as pivotal models to understand the mechanisms of the disease and evaluate potential treatments. These cell lines allow researchers to simulate biological processes in a controlled environment, enabling reproducibility and scalability. However, a major limitation of many studies is the lack of comprehensive demographic and genetic characterization of the cell lines used. Specifically, the underrepresentation or incomplete identification of ancestry information poses challenges to the generalizability and precision of research outcomes. Understanding the ancestry of cell lines is critical as it provides context for genetic variations, disease susceptibility, and drug response patterns among diverse populations. Addressing this gap is essential for achieving equitable and effective cancer therapies.
-
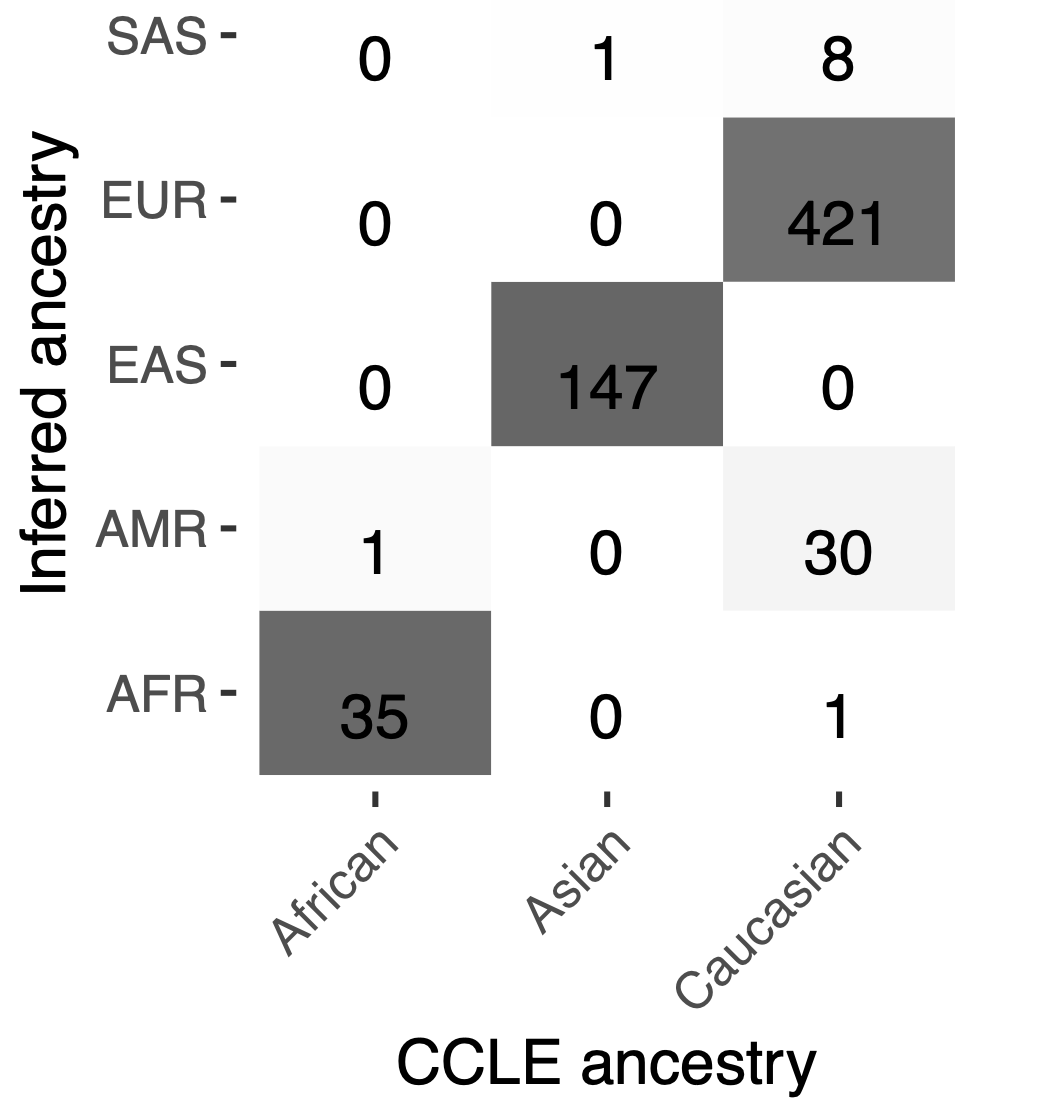
Bayesian inference pipeline to infer ancestry for cancer cell lines leveraging observed genotypes
My previous paper pointed out that ancestry is important for understanding the genetic diversity of cancer cell lines and their implications for therapeutic responses. This tutorial outlines the computational pipeline that I derived to reliably classify cancer cell lines into ancestries based on genotype data. The pipeline consists of three key stages: data processing, quality control, genotype imputation, and ancestry inference, each explained in detail to guide its implementation.
