Complex diseases present a significant challenge to modern medicine due to their multifactorial nature. Understanding these diseases requires the integration of diverse datasets that encompass clinical, environmental, genetic and metabolic factors. In recent years, together with the advances of molecular characterization technologies, the emergence of large multiomic and clinical datasets has opened new opportunities to study such diseases comprehensively. In this article, I discuss the challenges of stdudying complex diseases using multimodal datasets, our approach to tackle these challenges and an example study to demonstrate the effectiveness of our method.
Analysing multimodal datasets poses numerous challenges, such as high dimensionality, heterogeneity, and missing data. There are diverse sources of data, such as clinical measurements, imaging results, and omics data. Each data modality offers a unique perspective on the disease’s pathophysiology but also introduces specific challenges. The high dimensionality of omics data, for instance, can lead to overfitting in predictive models. Moreover, the heterogeneity of clinical and molecular data necessitates methods capable of reconciling diverse scales and structures. Missing data, a common issue in clinical context, further complicates the integration process. These challenges highlight the necessity for advanced analytical frameworks capable of leveraging the full potential of multimodal datasets while mitigating these issues.
IMML: A tool to integrate multimodal datasets to study complex diseases
In this study, I developed the Interpretable Multimodal Machine Leanrning (IMML) framework to address the aforementioned complications. The IMML framework is a clever two-step approach to effectively integrate heterogenous datasets and empower interpretability. The first step involves extracting functionally relevant features from each molecular layer independently, leveraging Gene Set Enrichment Analysis (GSEA). This approach ensured that each data modality’s unique contributions were captured effectively and projected to a common biological space. Subsequently, the second step benchmarked all possible combinations of data modalities using cross-validated forward feature selection and regularized linear models. The underlying hypothesis was that high-performing models utilizing the minimum number of data modalities would provide valuable insights into the etiology of the disease. An overview of the IMML framework is shown in Figure 1. Technical details of the method are described in a separate post in Projects.
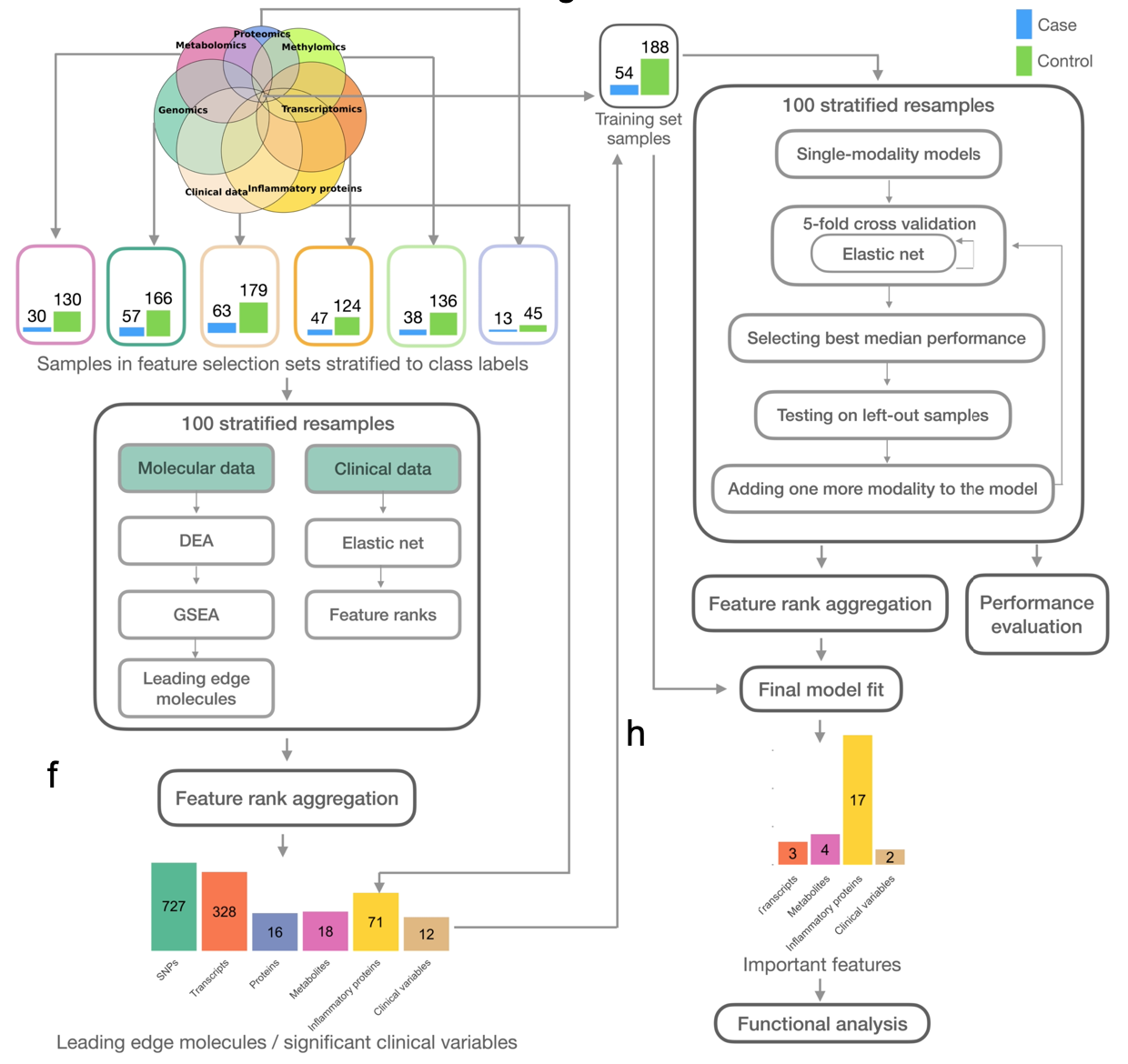
Figure 1. Overview of the IMML framework and example output.
IMML can stratify cross-sectional DSPN
Here I exemplified the performance of IMML leveraging a dataset of distal sensorimotor polyneuropathy (DSPN), a common neurological complication in diabetes, obesity patients and people with older age. Particularly, I aimed to stratify DSPN patients from healthy individuals (cross-sectional DSPN) and predict future DSPN outcome for healthy individuals (incident DSPN). The performance of the model was demonstrated by 100 resamplings of train and test sets from ~300 patients with available data modalities.
One advantage of IMML is that it could show which data modalities contribute the most to the prediction as it iterates through different model complexity and select the optimal combination of modalities to predict the target. In case of cross-sectional DSPN prediction, clinical data seemed to be the most influential modality to the prediction, as it was picked up in almost all the resamplings (Figure 2). When evaluating the prediction on the test set, we achieved high performance in many metrics, especially when only one modality (clinical data) was used (Figure 3). This highlighted the important of clinical data in cross-sectionally stratifying DSPN patients. Overall, IMML demonstrated good performance in feature selection and prediction for cross-sectional DSPN.
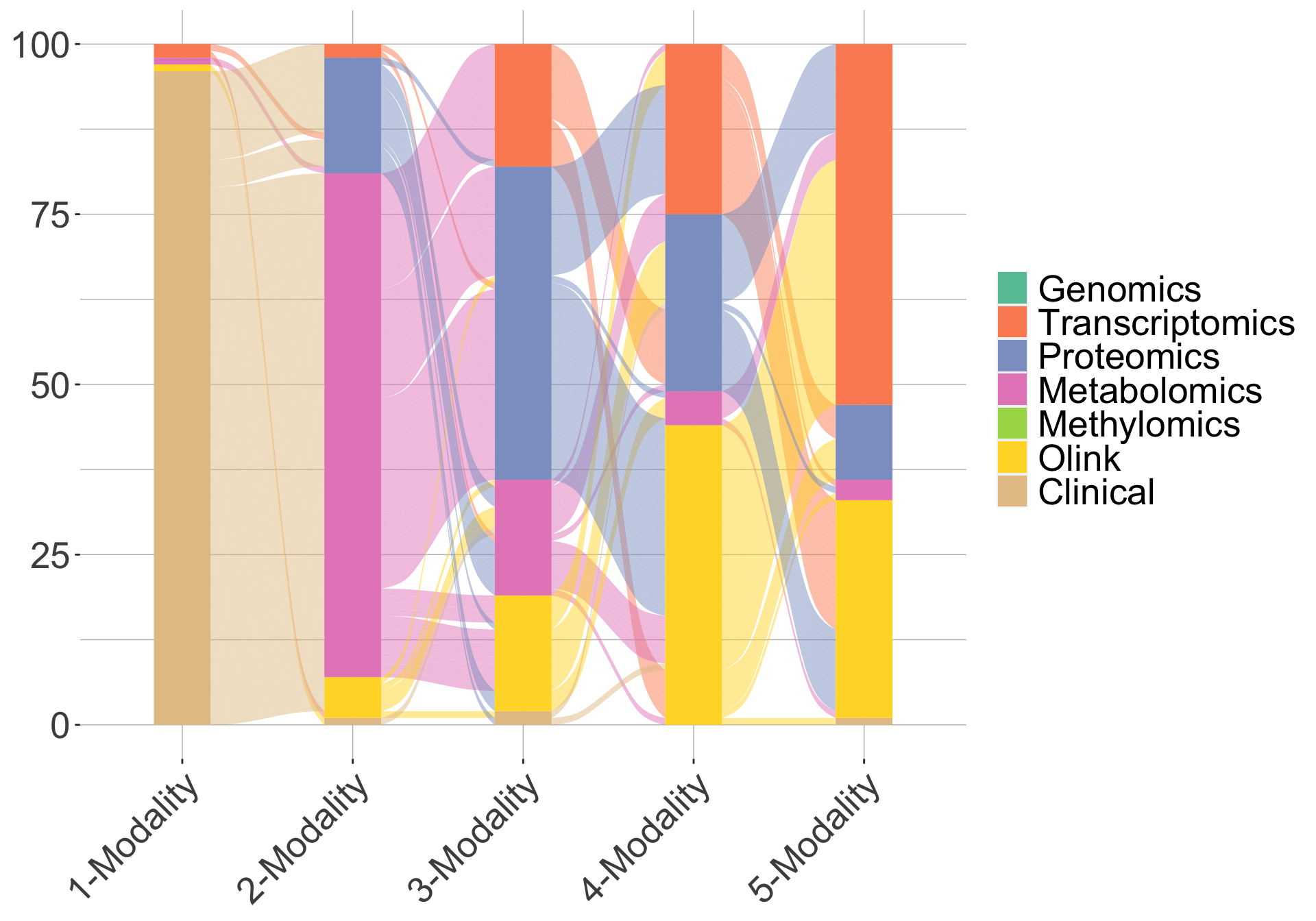
Figure 2. Trajectory of IMML's modality selection accross 100 resamplings for cross-sectional DSPN.One line represent one iteration in the 100 resamplings. Here only maximum 5 modalities were leveraged for the prediction.
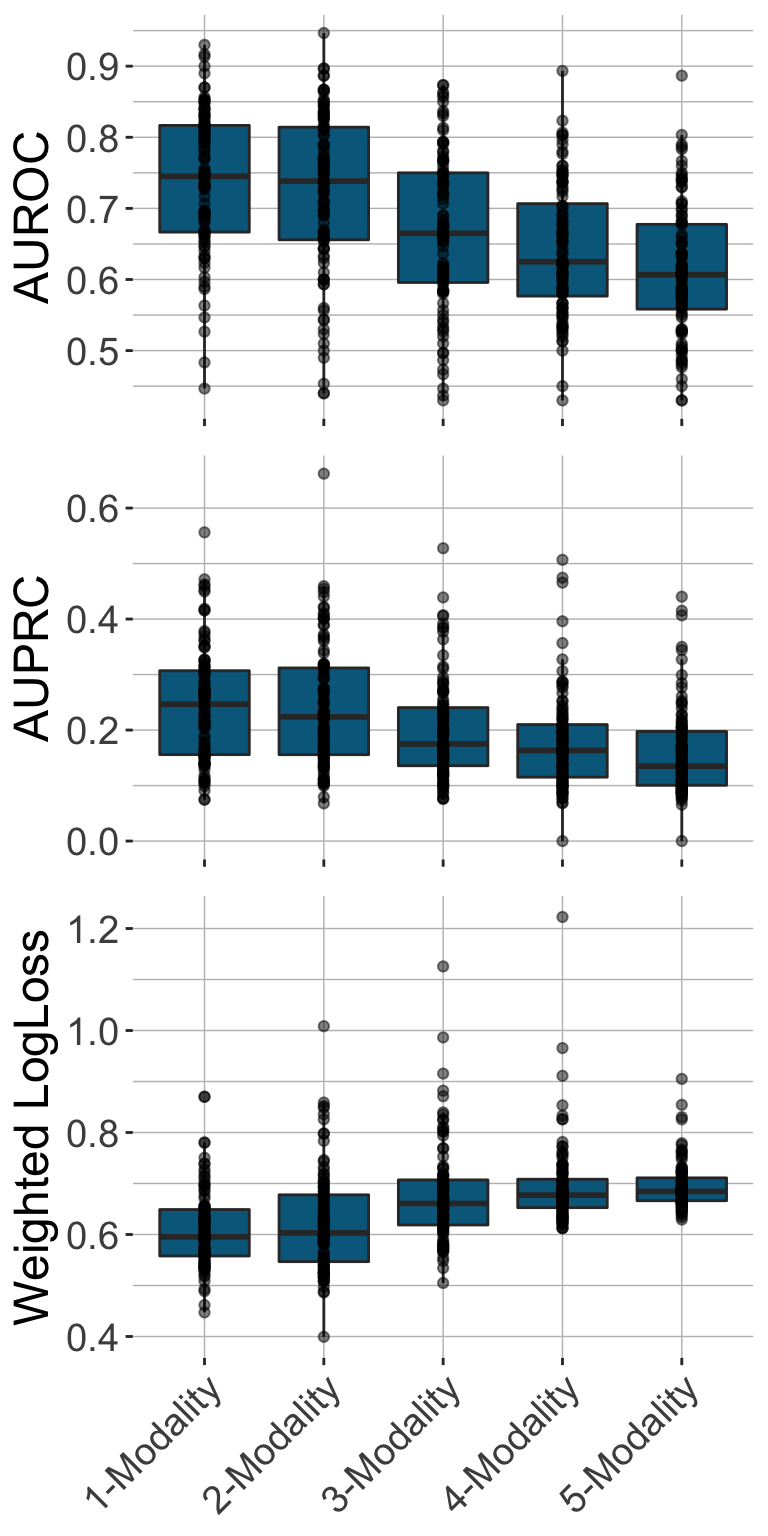
Figure 3. Performance of IMML in prediction of cross-sectional DSPN in the test sets. Here three metrics were used inlcuding AUROC, AUPRC and weighted log loss.
Results of Incident DSPN Prediction
In the context of incident DSPN, IMML also delivered robust predictions, and revealed insights into feature contribution. Specifically, the model showed more diverse feature contribution compared to the case of cross-sectional DSPN, as molecular data such as Olink protein panel, metabolomics and transcriptomics were picked up as the most dominating features (Figure 4). The model achieved maximum performance at 3-modality complexity (Figure 5). This underscored that multiomic data is necessary to study complex dieseases. In summary, IMML showed robust in prognosis of DSPN.
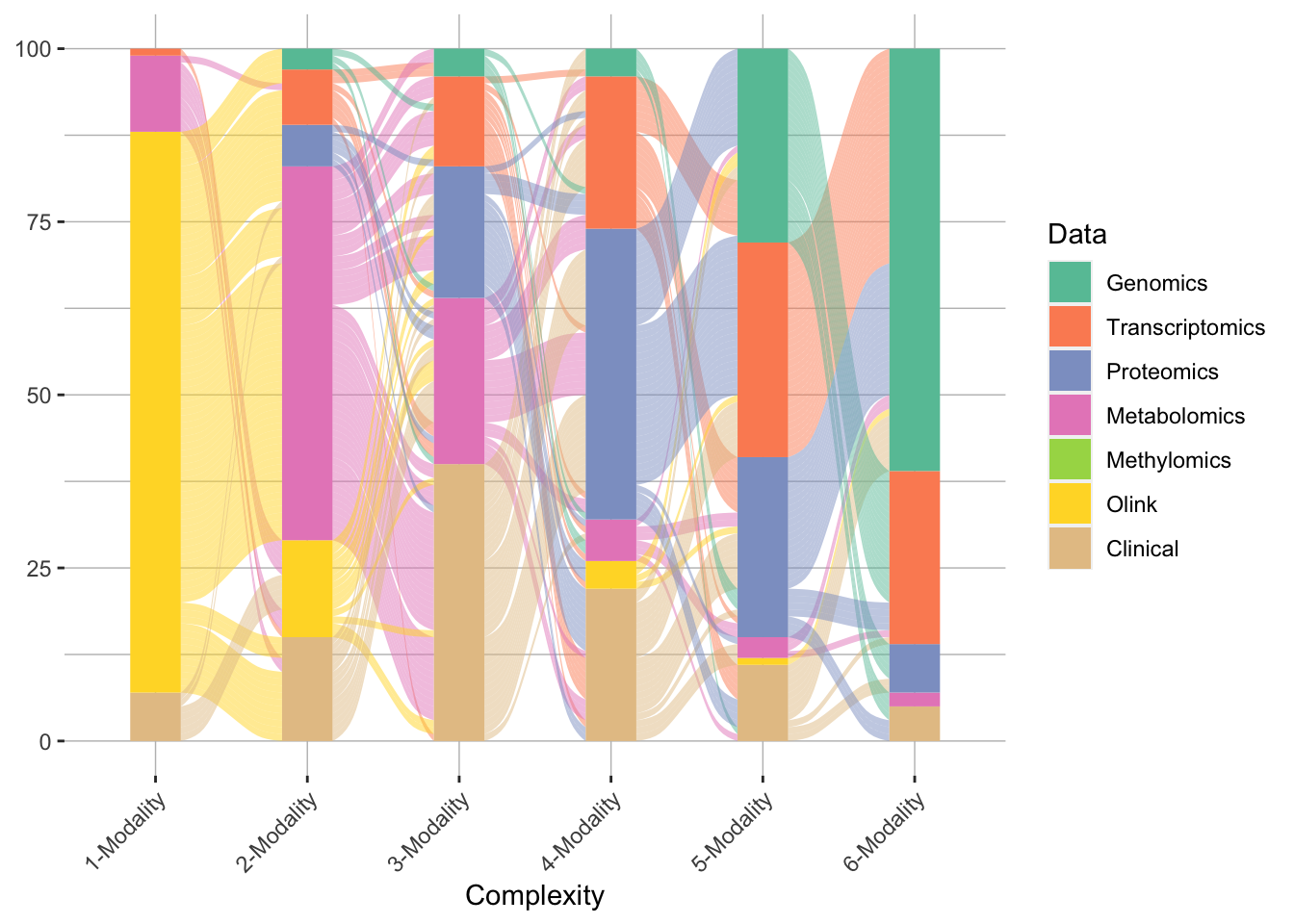
Figure 4. Trajectory of IMML's modality selection accross 100 resamplings for incident DSPN.One line represent one iteration in the 100 resamplings. Here maximum 6 modalities were leveraged for the prediction.
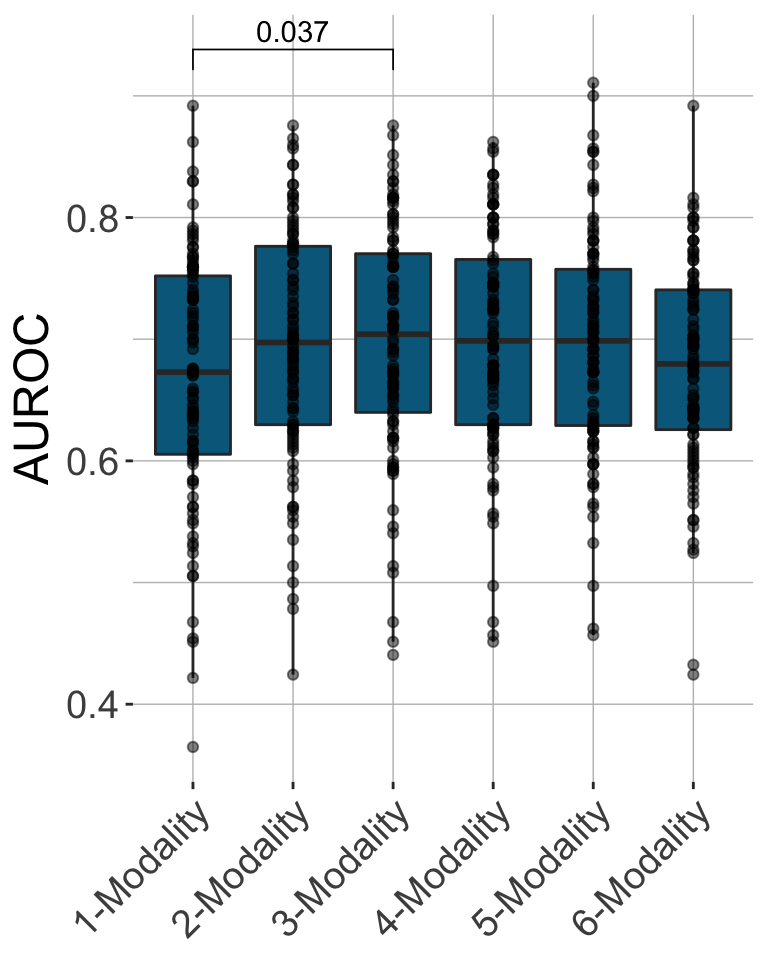
Figure 5. Performance of IMML in prediction of incident DSPN in the test sets.
Biomarkers of incident DSPN
One of the key advances of IMML is interpretability. By incorporating GSEA in the feature selection process, I made it straight forward to select biologically relevant features that were predictive for the phenotype of interest. In case of incident DSPN prediction, analysis of features revealed multiple important predictors for the phenotype. Among the key features detected were metabolic markers such as elevated levels of inflammatory proteins and essential fatty acids, which were consistently linked to disease onset. Decreased levels of SUMOylation transcripts also contributed to the prediction significantly (Figure 6). Furthermore, factors such as age and physical activity levels emerged as critical contributors. These findings underscore the utility of IMML in uncovering novel, actionable insights into disease mechanisms and empowering putative biomarker discovery.
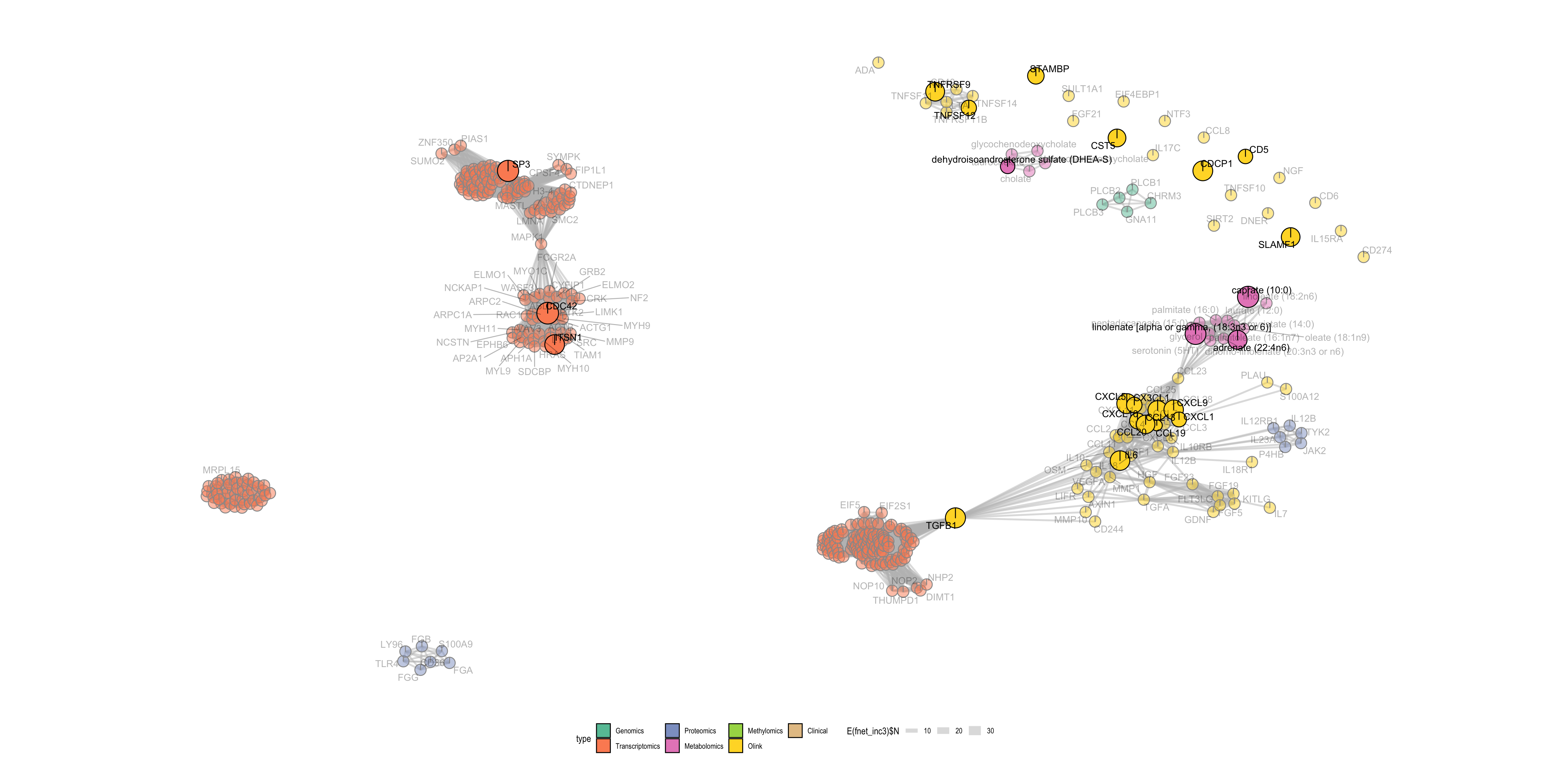
Figure 6. Important molecular features for incident DSPN prediction. Nodes are features and edges are gene sets (in the GSEA) that these features are involved.
Take home message
Here are a few key points to summarise the paper:
- IMML is a novel machine learning framework to select and integrate multiomic data for phenotype prediction leverging prior biological knowledge through GSEA
- IMML demonstrated robust performance in studying DSPN, both achieving high prediction accuracy and discovery of relevant and actionable biomarkers for the disease.
Details can be found in the publsihed paper. Analysis code and framework is in the github repository.
