In the previous post we got to know the IMML and its application in the DSPN study. In this post I would like to go more into details of the algorithm, how it is constructed, what it actually does underneath and how it helps in studying complex deseases. This is more of a discription of the algorithm, rather than a coding tutorial. For a full on tutorial of how to actually use IMML, please visit my github repository.
Components of IMML
IMML is an end-to-end machine learning framework for prediction of complex diseases’ phenotypes and discovery of biomarkers by integrating multimodal datasets. To overcome the technical challenges of analyzing vast and heterogenous biomedical datasets, IMML implements a multistep approach from data preparation to feature selection and integration. Specifically, IMML framework consists of four consecutive components: data partitioning, feature selection, model training and feature analysis. The four components resembles a standard machine learning workflow, but were customised to addess the aforementioned challenges and maximize performance and interpretability. Figure 1 gives an overview of the framework including the components and their input/output. Subsequent chapters will dive into details of these components.
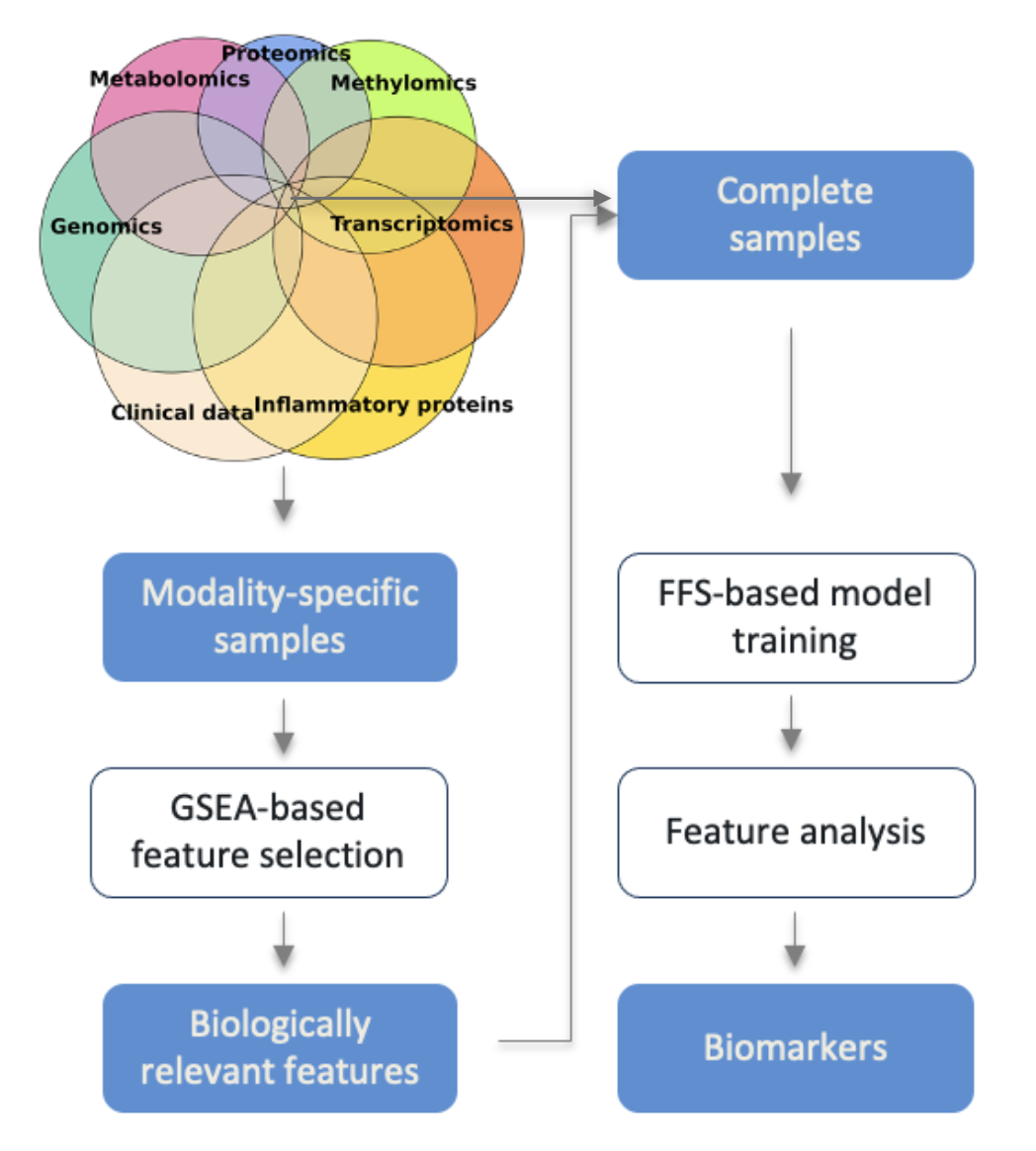
Figure 1. Overview of the IMML framework and its components.
Data partitioning
The first step in the framework is data partitioning, which was derived to overcome the issue of missing data and prepare the input for subsequent feature selection and model training. Missing data is a quite common problem in biomedical datasets, as not all patients were characterized with the same procedure and data originates from various sources, leading to an uneven distribution of features among individuals. An example in Figure 2 shows that combinations of data modalities from the same dataset have varying sample sizes.
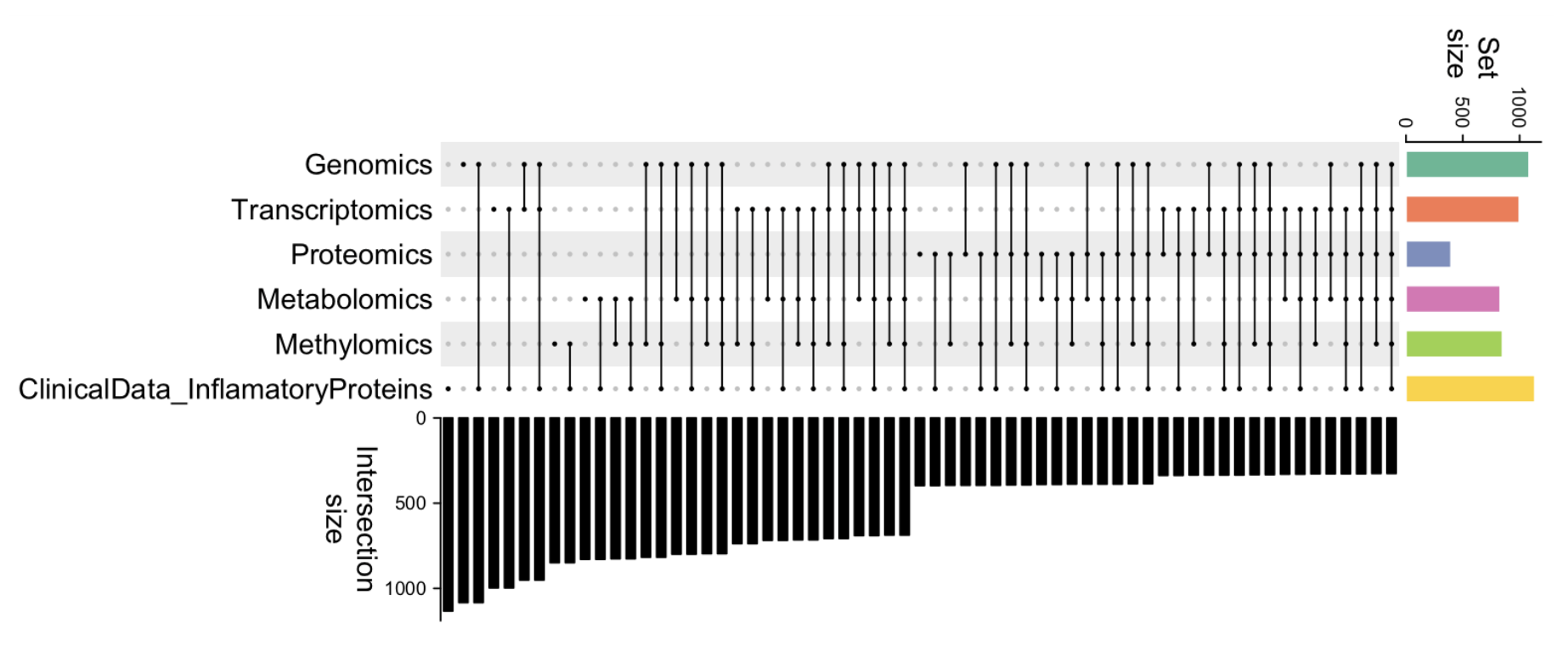
Figure 2. Example of a dataset with uneven distribution of samples in different data modalities.
To maximize the utility of data, I split the dataset into two parts. The subset of samples that are fully characterized, ie. having all features, is set aside for model training later on. For the rest of the samples, samples that are fully characterized for each data modality are put into separate subsets for feature selection. The feature selection process is done separately for each modality and utilize the corresponding subsets. An example is shown in Figure 3, where the data is split into one model training set and several feature selection sets. Using this approach, we tackle the issue of uneven feature distribution and improve the robustness of feature selection and model training.
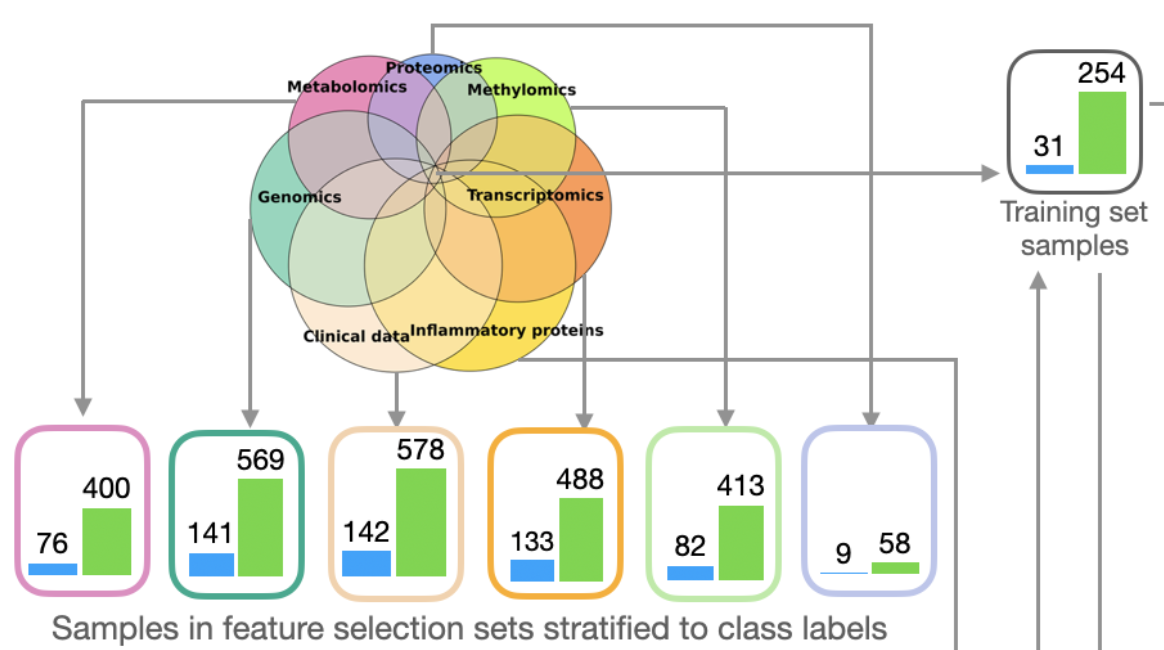
Figure 3. Example of the data partitioning output. Blue bars show positive (case) samples and green bars are negative (control) samples.
Feature selection
One of the advances of IMML lies in the feature selection process. This step aims to shrink the vast and heterogenous feature space of the analyzed dataset into an ML-manageable and interpretable space. To this end, I leverage gene set enrichment analysis (GSEA) to select the features that are both predictive for the phenotype and biologically relevant to incorporate into the model. Particularly for each molecular modality, GSEA is performed to identify the significantly enriched signaling pathways associated with the phenotype and the specific set of features that drive the enrichment of these pathways (leading edge features). Please refer to the GSEA publication for details of the algorithm and how to identify leading egde features. In short, GSEA starts with a ranked feature list (ranked from the most up-regulated to the most down-regulated in association with the phenotype) and statistically evaluates whether a predefined feature set (corresponding to a signaling pathway) significantly locates toward the top (up-regulated) or bottom (down-regulated) of the feature list, through hypothesis testing of an enrichment distribution against a null distribution of data generated by permutation. Here the GSEA is conducted separately for each data modality using the previously prepared feature selection sets and the selected leading edge features are combined at the end to train the model in the subsequent model training step. Figure 4 gives an overview of input and output of the feature selection. Altogether, this approach ensures that the originally heterogenous feature space is projected to a smaller common biological space to boost predictablity and interpretability.
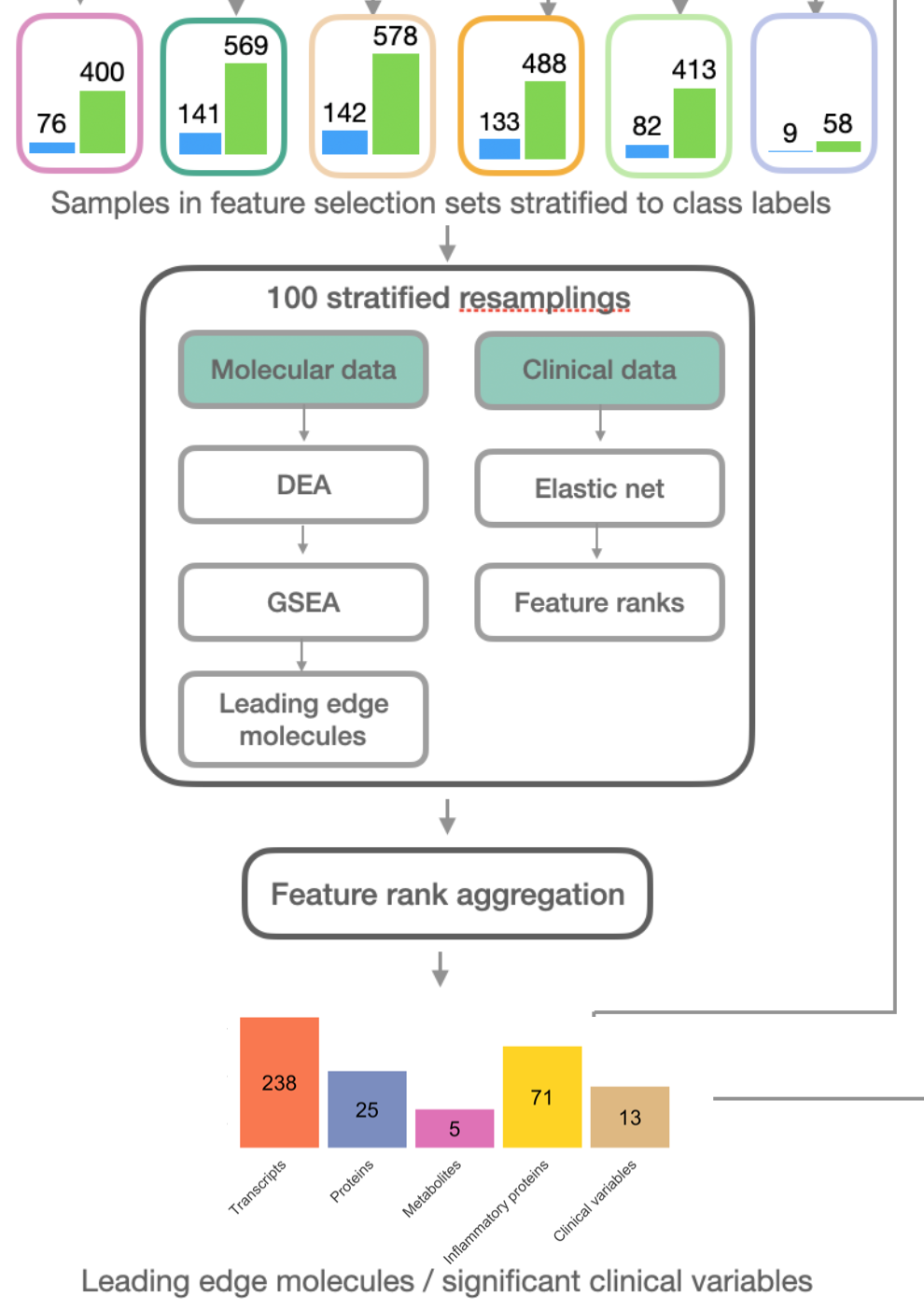
Figure 4. Feature selection procedure of IMML Each modality is selected separately through GSEA and the leading edge features are combined together at the end.
Model training
In this final step IMML leverages the fully characterized samples the I set aside in the data partitioning step together with the leading edge features selected in the feature selection step to make up a dataset for training. To this end IMML makes 100 stratified splits, each of which allocates 80% of the data for feature integration and training, with 20% reserved for testing. The training data was further divided into stratified 5-fold cross-validation sets. Cross-validation results guided the iterative forward feature selection (FFS) algorithm, which incrementally integrates multiple data modalities by evaluating performance of modality combinations over the 5-fold cross-validation. The FFS starts with a single modality and selects more modalities to add to the model through cross-validation until all features are included. To assist interpretability, by default IMML uses elastic net as the training algorithm, but in theory one can choose between algorithms such as Random Forest, Support Vector Machine and XGBoost. IMML also has many flavors of performance metrics, including AUROC, AUPRC and Weighted Log Loss. The choice depends on data situations. After training IMML aggregates important features (based on the t-statistics of the linear models’ parameters) from the 100 runs using Robust Rank Aggregation (RRA). These features were used to train a final elastic net model on the entire training dataset, with feature importance evaluated through t-statistics of the model parameters. Figure 5 gives an overview of the model training and important feature extraction. In summary, the model training process the final step to integrate multimodal features, make prediction and evaluate biomakers of the phenotype.
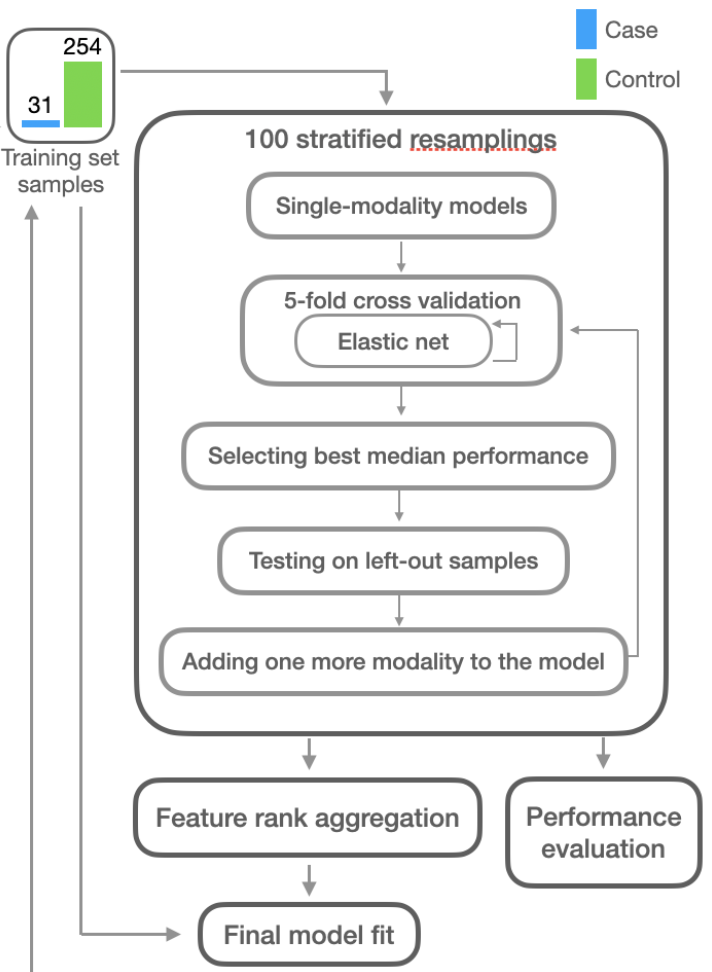
Figure 5. Model training and important feature extraction
Take home message
-
IMML is an end-to-end machine learning framework to make prediction and discover biomarkers of complex diseases, utilising GSEA and robust model training algorithm
-
Full instruction how to install and use the tool can be found in the github repository. Also refer to the paper for detailed description of the algorithm and its application.
