A bit about Interpretability
15 Mar 2025
Interpretability in machine learning is one of the topics I paid attention to throughout my PhD. Especially in biomedical research, building models that predict outcomes accurately is thrilling, but it’s not the entire story. The magic happens when we truly understand what these models tell us and translate those insights into something meaningful. In this post, let’s dive into why interpretability matters and explore some practical ways to make our models both understandable and impactful.
1. Interpretability in Biomedical research
Interpretability isn’t just a fancy word—it’s crucial for translating ML models into real-world biomedical applications. It helps scientists uncover biomarkers, provides deeper mechanistic insights into diseases, and assists biotech and pharma in drug discovery. Equally important, interpretability boosts trust: doctors, researchers, and even patients can better accept and use ML tools when they understand what’s behind predictions.
Simpler models often provide better interpretability. For instance, let’s take a look at this cute picture comparing a decision tree and a deep neural network:
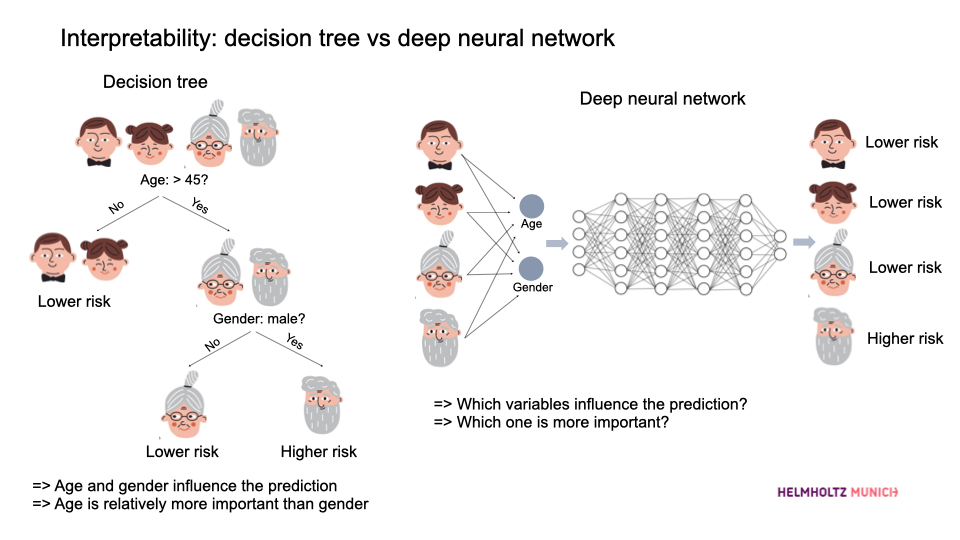
Comparison between decision tree and deep neural network.
In the decision tree, we see exactly how decisions are made (e.g., age and gender directly influence the risk, and age is relatively more important). It’s as clear as preparing an organic fruit salad—you see exactly what’s going in, fresh and straightforward! Conversely, the deep neural network is more like a premade smoothie—delicious but with mysterious ingredients blended together, leaving you guessing what’s inside!
2. Inherently interpretable algorithms
In biomedical research, we often prefer simpler models such as linear regression, logistic regression, or decision trees. Why? Because most biomedical studies typically have smaller sample sizes and limited computational resources. Besides, simpler models generally perform reasonably well for tasks like classification and forecasting.
These simpler models come with inherent interpretability, meaning you can directly extract feature importance from the model parameters themselves. For example:
-
Linear models: The coefficients directly indicate the importance and direction (positive or negative influence) of each feature. Like picking out individual fruits, you clearly see what and how much contributes to your tasty salad.
-
Tree-based models (Decision Trees, Random Forests): You can explicitly visualize and interpret how each feature influences predictions through tree splits. Feature importance can be quantified by how much each feature reduces uncertainty or error—each decision step.
Thus, choosing simpler algorithms often gives you immediate interpretability, making your life easier—no detective work needed here, just fresh ingredients!
3. Posthoc interpretability in deep learning
But what if we have sufficient samples, resources, and a strong desire to push predictive power to the limits? Deep learning is our superhero, though it usually comes wrapped in a complex, mysterious package—much like a premade smoothie. Tasty? Yes. Transparent? Not always!
Fortunately, we have posthoc methods to make sense of these complex models after they’ve been trained:
-
SHAP (SHapley Additive exPlanations) : SHAP assigns each feature an importance value for a particular prediction, inspired by cooperative game theory. It explains individual predictions clearly by measuring each feature’s contribution.
-
Integrated Gradients : Integrated gradients measure the contribution of each input feature by integrating gradients along the path from a baseline input (often zero) to the actual input. It reveals how changing each input slightly affects the model’s prediction.
These methods act like nutritional labels or ingredient lists—turning your mysterious smoothie into something you can confidently savor!
4. Incorporation of biological knowledge
Adding biological expertise is like sprinkling some expert seasoning that you know exactly what it does onto your dish—it elevates the entire analysis. By integrating prior biological knowledge during feature selection, we do not only narrow down the search space to “relevant features”, but also enhance interpretability by associating our features with prior biological knowledge that might boost mechanistic explaination. Resources like databases for molecular attributes and phenotype associations (e.g., KEGG, Reactome, Gene Ontology) significantly empower our interpretations.
However, be cautious here, biological knowledge is limited. The current available knowledge might not be sufficient to explain all the variance within our examined dataset. Always pair it with unbiased approaches to ensure comprehensive and balanced analyses.
Final remarks
Interpretability isn’t just icing on the ML cake—it’s the flour holding it all together. By prioritizing understandable models and cleverly combining computational methods with biological knowledge, we can unlock the true potential of ML in biomedical research.
Above are some thoughts for building ML models we can trust, understand, and genuinely enjoy unraveling!
Happy interpreting!
